Descripciones de casos
Dog registration hanging on a tooth: a case report
Registro de perros a partir de un diente: descripción de un caso
Analecta Veterinaria
Universidad Nacional de La Plata, Argentina
ISSN: 0365-5148
ISSN-e: 1514-2590
Periodicity: Frecuencia continua
vol. 44, e088, 2024
Received: 17 May 2024
Revised: 13 August 2024
Accepted: 20 August 2024
Corresponding author: guillermogiovambattista@gmail.com

This work is licensed under Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International.
Abstract: Dog breeder associations have regulations for the registration and breeding of their individuals. The complete dentition is between them, thus the absence of first and second molars prevents the registration of specimens. A dispute arose when the registration of a 20-month-old male dog was rejected by the Argentine club of German Shepherd Dog Breeders, because the second lower right molar was absent in routine examination. The breeder appealed the decision, alleging that the absence of the molar was accidental and not for a genetic reason, providing as evidence a molar found in his breeding kennel. Our laboratory was consulted with the objective of determining the genetic relationship between the oral swab sample obtained from the rejected dog (reference), and the molar found (evidence). For this reason, genotyping was carried out through the use of microsatellites. The results confirmed that the samples shared the same DNA profile, with the molar found being between 1.00E+11 and 1.27E+17 times more likely to have come from the dog to be registered than from an animal taken at random from the reference population. To date, there are no known reports of a similar dispute, resolved by studying a DNA profile using a tooth.
Keywords: Registration dispute, German Shepherd dog, molar tooth, microsatellites, genetic identification.
Resumen: Las asociaciones de criadores de distintas razas caninas tienen reglamentaciones para la inscripción y cría de sus individuos. La dentición completa es una de ellas, por lo que la ausencia de primeros y segundos molares impide el registro de ejemplares. Se presentó una disputa en la inscripción de un perro macho de 20 meses de edad que fue rechazada por el Club Argentino de Criadores del Perro Ovejero Alemán, debido a la ausencia del segundo molar inferior derecho. El criador apeló la decisión alegando que esta ausencia fue accidental y no por una razón genética, aportando como evidencia un molar encontrado en su criadero. Se consultó a nuestro laboratorio con el objetivo de determinar la relación genética entre la muestra de hisopado oral obtenida del animal a inscribir (referencia) y la del molar encontrado (evidencia). La genotipificación mediante el uso de microsatélites confirmó que ambas muestras compartían el mismo perfil de ADN, siendo entre 1,00E+11 y 1,27E+17 veces más probable que el molar encontrado procediera del perro a registrar que de un animal tomado al azar de la población de referencia. Hasta el momento no se conocen comunicaciones de una disputa similar, resuelta mediante el estudio de un perfil de ADN utilizando una pieza dentaria.
Palabras clave: Registro de perros, Ovejero alemán, molar, microsatélites, identificación genética.
Introduction
Non-human forensic genetics is the application of genetics to non-human material for the resolution and prevention of legal conflicts (Kanthaswamy, 2015). The first case reports were published about two decades ago (Giovambattista et al., 2001; Savolainen & Lundeberg, 1999; Schneider et al., 1999). At the beginning, this discipline was applied to the investigation of crimes inherent to human beings using DNA evidence from animals, plants, bacteria and viruses (Barrientos et al., 2016; Kanthaswamy, 2015). However, non-human forensic genetics has grown significantly in the last few years, extending its application to the analysis of cases of animal theft, contamination/adulteration, illegal traffic/poaching, animal attack, animal cruelty and horse doping, among others (Díaz et al., 2008; Di Rocco et al., 2011; Kanthaswamy et al., 2021).
Argentina is the country with the highest number of pets per capita in Latin America, with around 9 million dogs and 3 million cats, without counting stray dogs. Each year, more than 1,000 purebred dogs are exported to around 46 countries (SENASA, 2018, http://web.senasa.gob.ar), making pet breeding an important activity in Argentina. To maintain the quality of the animals bred and guarantee quality breeding to domestic and foreign buyers, the different breeder clubs have determined strict rules for the registration of animals in herd books.
The breeding and registration regulations of the Argentine club of German Shepherd Dog Breeders (Club Argentino de Criadores del Perro Ovejero Alemán, POA by its acronym in Spanish, https://clubpoa.com.ar/reglamentos/crianza-y-registro) determine that the use of dogs with absence of a first or second molar is prohibited for breeding. In addition, these animals are severely penalized in breed dog shows. Here we report a new application of non-human forensic genetics, which was used to solve a registration dispute and prevent a civil litigation.
Case report
Description
A German Shepherd breeder failed to register a 20-month-old male dog in POA because the lower right second molar of animal was absent when routine examination was carried out. The dog breeder appealed the decision, alleging that the absence of this molar was caused by an accident and not due to a genetic reason. As evidence, the breeder provided a lower right second molar tooth found in the breeding kennel. Our laboratory was consulted with the objective of determining the genetic relationship between the oral swab sample obtained from the rejected dog (reference) and the molar found (evidence).
Biological samples
Two samples were anaysed: one oral swab obtained from the German Shepherd dog (reference), and one tooth sample (evidence) provided by the breeder to demonstrate that the dog had complete dentition. Both samples were kindly provided by POA and received and analysed at the laboratory (IGEVET, UNLP-CONICET LA PLATA). POA required the owner of the animal to provide consent to report the present case, before sending the sample to IGEVET.
DNA extraction and genotyping
Before DNA extraction, the tooth was subjected to a decalcification process using a 0.5 M ethylenediaminetetraacetic acid (EDTA) - 20% N-lauroylsarcosine solution. Then, genomic DNA was purified using an in-house organic extraction method. The DNA from the oral swab sample was obtained using an extraction buffer containing 100 mM tris-hydrochloride pH 8, 10 mM EDTA, 2 M NaCl, 10% sodium dodecyl sulfate (SDS), and 10 ng/µl proteinase K. The DNA quantity and quality were measured using the Multiskan™ GO Microplate Spectrophotometer (Thermo Fisher Scientific, USA) and electrophoresis in 1% - 0.5 X tris, borate and EDTA (TBE) agarose gels.To determine if both reference and evidence samples belonged to the same dog, the obtained DNA were genotyped using the Canine Genotypes Panel 1.1 kit (Thermo Fisher Scientific) that encompasses the following 18 autosomal microsatellites: AHTk211, CXX279, REN169O18, INU055, REN54P11, INRA21, AHT137, REN169D01, AHTh260, AHTk253, INU005, INU030, FH2848, AHT121, FH2054, REN162C04, AHTh171, REN247M23, and a sex-determining locus (amelogenin). These markers are included in the 'core panel' of loci recommended by the Applied Genetics Committee of Companying Animals of the International Society for Animal Genetics (ISAG, http://www.isag.org.uk) for canine genetic identification and parentage analyses. Fragments were resolved in an Applied Biosystems 3500 sequencer and analyzed using GeneMapper®v4.1 (Thermo Fisher Scientific).
Statistical analyses
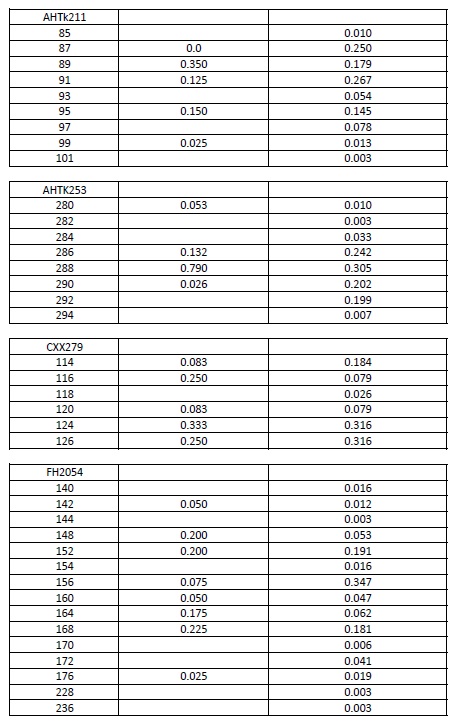
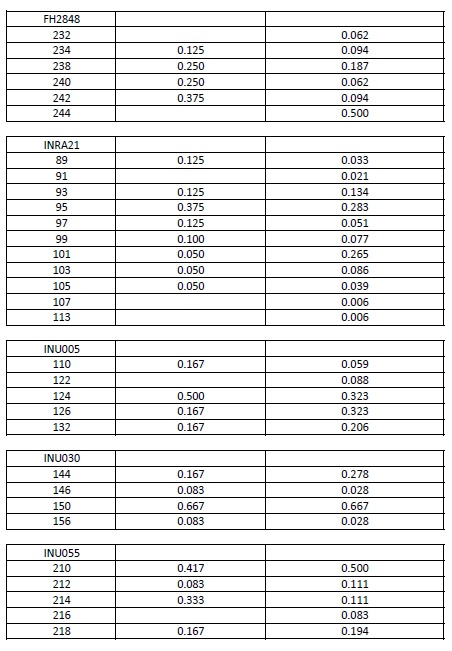
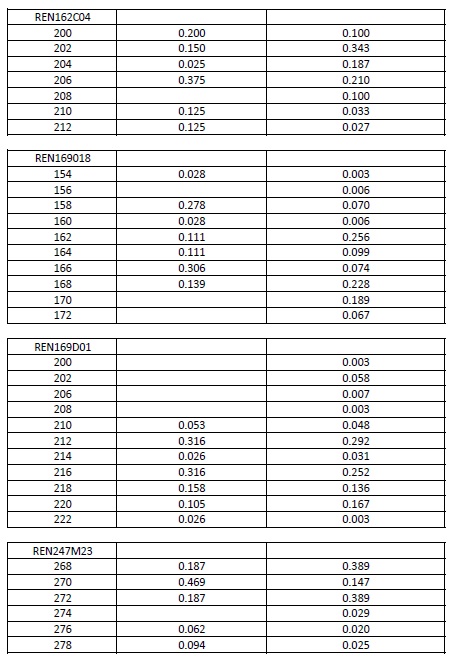
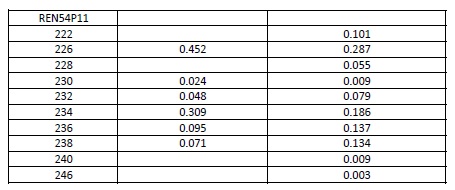
Allele frequencies of each microsatellite were calculated using the genotypes of the IGEVET database, which comprises 169 dogs from 14 breeds (including 21 German Shepherd dogs). The likelihood ratio (LR) and the random-match probability (MP) between the DNA profiles of the evidence and reference samples were calculated for each STR and for the whole set using the equations proposed by Balding & Donnelly (1995). For these estimations, 0.27 and 0.09 . values among breeds were used (Kanthaswamy et al., 2009; Parker et al., 2004). Furthermore, these parameters were calculated considering that the molar tooth found could belong to a relative of the declared dog, including the relatedness coefficients (k., 2k., and k.) into the formula (Weir, 2003). For these cases, 0.019 . value among kennel was used (Crespi et al., 2018). All data are provided either in the text or as Supplementary material (Table S1).
Results and discussion
Comparison of the genotypes obtained from the molar and the oral swab evidenced that both samples had the same DNA profile (Figure 1). Thus, the declared dog cannot be excluded as the source of the molar tooth found in the kennel.

Figure 1
Electropherogram of the molar tooth found in the kennel (a) and the oral swab of the declared German Shepherd dog (b). A sector in which the fluorochromes used are identified (A: black, B: red, C: blue and D: green) is highlighted with a red circle, allowing the agreement between both samples to be observed.
In addition, the amelogenin marker confirmed the female gender of the evidence sample. The LR showed that it was between 1.00E+11 and 1.27E+17 times as likely that the tooth came from the declared German Shepherd dog than from a random unrelated animal from the dog population. Usually, relatives of dogs from different generations and litters live in a kennel (e.g., half or full siblings, parents-offspring, first cousins, uncle-nephew, and grandparent-grandchild) because dogs begin their reproductive activity very young. For this reason, a close relative of the declared dog could be the source of the tooth (Table 1).
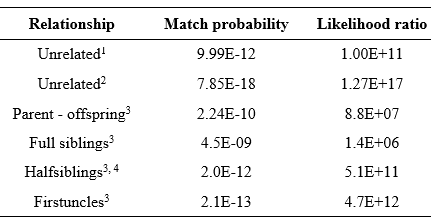
Estimated values using 1a θ value of 0.27 among breeds, 2a θ value of 0.09 among breeds, 3a θ value of 0.019 among kennels. 4Uncle-nephew and grandparent-grandchild had the same match probability.
As expected, these analyses showed lower values of match probability in comparison with scenarios of unrelated dog sources. However, even in the worst scenario (the source of evidence came from a full sibling) the obtained match probability would be enough with reference to the population size. These results agree with those reported by Arizmendi et al. (2020) in breeding kennels of Doberman Pinscher dogs, showing that the standard panels had enough discrimination power for genetic identification within a kennel.
Conclusion
To the best of our knowledge, this is the first report of a dog registration dispute between a breeder and a dog club that was solved using STR DNA profiling of a tooth,thus expanding the types of application of non-human forensic genetics.
Acknowledgements
We thank the Argentine club of German Shepherd Dog Breeders for providing the samples and A. Di Maggio for careful manuscript correction.
Funding
This study was supported by the National University of La Plata (Grant V247).
Conflict of interest
The authors declare that they have no conflict of interest.
Authorship statement:
Conceived the project and designed the study: JAC, EEVC, and GG. Sample collection and data acquisition: JAC, EEVC, AA, NSC, and MEZ. Analysed the data: JAC; EEVC; AA; NSC; MEZ, and MEF. Contributed to reagents/materials/analysis tools acquisition: GG, and PPG. Drafted and revised the manuscript: JAC, EEVC, NSC, MEZ, and GG. All authors have read and agreed to the published version of the manuscript.
References
Arizmendi A, Batista PR, Crespi JA, Tórtora M, Vercellini M, Czernigow M, Arias DO, Giovambattista G. 2020. Analysis of PDK4 gene deletion in a population of Doberman Pinschers from Argentina. Research communications of the 30th ecvim-ca online congress. Journal of Veterinary Internal Medicine. 34 (6):3058-3166. https://doi.org/10.1111/jvim.15924
Balding DJ, Donnelly P. 1995. Inference in forensic identification. Journal of the Royal Statistical Society. Series A (Statistics in Society). 158 (1): 21-53. https://doi.org/10.2307/2983402
Barrientos LS, CrespiJA, FameliA, PosikDM, MoralesH, Peral García P, GiovambattistaG. 2016. DNA profile of dog feces as evidence to solve a homicide. Legal Medicine (Tokyo, Japan). 22:54-7. https://doi.org/10.1016/j.legalmed.2016.08.002
Crespi JA, Barrientos LS, Giovambattista G. 2018. von Willebrand disease type 1 in Doberman Pinscher dogs: genotyping and prevalence of the mutation in Buenos Aires region, Argentina. Journal of Veterinary Diagnostic Investigation. 30(2):310-4. https://doi.org/10.1177/1040638717750429
Díaz S, Kienast ME, Villegas-Castagnasso EE, Pena NL, Manganare MM, Posik D, Peral García P,Giovambattista G. 2008. Substitution of human for horse urine disproves an accusation of doping. Journal of Forensic Sciences. 53(5):1145-8. https://doi.org/10.1111/j.1556-4029.2008.00797.x
Di Rocco F, Posik DM, Ripoli MV, Díaz S, Maté ML, Giovambattista G, Vidal Rioja L. 2011. South American camelid illegal traffic detection by means of molecular markers. LegMed (Tokyo). 13(6):289-92. https://doi.org/10.1016/j.legalmed.2011.08.001
Giovambattista G, Ripoli MV, Lirón JP, Villegas Castagnasso EE, Peral García P, Lojo MM. 2001. DNA typing in a cattle stealing case. Journal of Forensic Sciences. 46(6):1484-6.
Kanthaswamy S. 2015. Review: domestic animal forensic genetics-biological evidence, genetic markers, analytical approaches and challenges. Animal Genetics. 46(5):473-84. https://doi.org/10.1111/age.12335
Kanthaswamy S, Brendel T, Cancela L, Andradre de Oliveira DA, Brenig B, Cons C, Crespi JA, Dajbychová M, Feldl A, Itoh T, Landi V, Martínez A, Natonek-Wisniewska M, Oldt RF, Radko A, Ramírez O, Rodellar C, Ruiz-Girón M, Schikorsk D, Turba ME, Giovambattista G. 2021. An inter-laboratory study of DNA-based identity, parentage and species testing in animal forensic genetics. Forensic Sciences Research.7(4):708-13. https://doi.org/10.1080/20961790.2021.1886679
Kanthaswamy S, Tom BK, Mattila AM, Johnston E, Dayton M, Kinaga J, Erickson BJ, Halverson J, Fantin D, DeNise S, Kou A, Malladi V, Satkoski J, Budowle B, Smith DG, Koskinen MT. 2009. Canine population data generated from a multiplex STR kit for use in forensic casework. Journal of Forensic Sciences. 54(4):829-40. https://doi.org/10.1111/j.1556-4029.2009.01080.x
Parker HG, Kim LV, Sutter NB, Carlson S, Lorentzen TD, Malek TB, Johnson GS, DeFrance HB, Ostrander EA, Kruglyak L. 2004. Genetic structure of the purebred domestic dog. Science. 304(5674):1160-4. https://doi.org/10.1126/science.1097406
Savolainen P, Lundeberg J. 1999. Forensic evidence based on mtDNA from dog and wolf hairs. Journal of Forensic Sciences. 44(1):77-81.
Schneider PM, Seo Y, Rittner C. 1999. Forensic mtDNA hair analysis excludes a dog from having caused a traffic accident. International Journal of Legal Medicine. 112(5):315-6. http://doi.org/10.1007/s004140050257
Weir BS. Forensics. In: Balding DJ, Bishop M, Cannings C. 2008. In: Handbook of Statistical Genetics. 3rd edition. Hoboken, New Jersey. John Wiley & Sons, pp 1368-92.
Supplementary material

Author notes
Correo electrónico del autor de contacto: guillermogiovambattista@gmail.com


